The September 2009 issue of Nature Biotechnology has a letter concerning the receptor specificity of AH1N1 2009 pandemic influenza virus – which accounts pretty well for why it CAN be pretty nasty, and for why it may get nastier yet.
Childs et al., in a letter entitled “Receptor-binding specificity of pandemic influenza A (H1N1) 2009 virus determined by carbohydrate microarray“, describe what amounts to a tour de force analysis of the receptor binding of a number of influenza viruses, which concludes with the statement that:
“The differences in receptor binding between the 2009 pandemic and seasonal H1N1 viruses may therefore account, at least in part, for the higher virus replication and greater pathology reported in the lungs of ferrets, mice and nonhuman primates infected with pandemic viruses, than observed with contemporary seasonal viruses.”
Which would help explain why some otherwise healthy young people are dying of the virus, while others are getting only mildly ill. But we get ahead of ourselves: in January last year I wrote in MicrobiologyBytes about recpetor specificities of A-type influenza viruses, in the context of how H5N1 was less likely to mutate to easy human-to-human transmissibility than had origianlly been thought.
I wrote at the time:
According to a letter in the January 2008 issue of Nature Biotechnology, it is a characteristic structural topology, and not just the α2,6 linkage, that enables specific binding of HA to α2,6 sialylated glycans. The authors state:
…recognition of this topology may be critical for adaptation of HA to bind glycans in the upper respiratory tract of humans. An integrated biochemical, analytical and data mining approach demonstrates that HAs from the human-adapted H1N1 and H3N2 viruses, but not H5N1 (bird flu) viruses, specifically bind to long α2-6 sialylated glycans with this topology. This could explain why H5N1 viruses have not yet gained a foothold in the human population.
Apparently the critical shape in humans is umbrella-like, whereas the avian receptor is characteristically cone-like. Again from the paper:
The topology of α2-3 and α2-6 is governed by the glycosidic torsion angles of the trisaccharide motifs-Neu5Aca2-3Galb1-3/4GlcNAc and Neu5Aca2-6Galb1-4GlcNAc, respectively (Supplementary Fig. 3 online).
Ram Sasisekharan and colleagues showed that human-adapted viruses with mixed α2,3/α2,6 binding ability that bound the umbrella-type receptor were efficiently transmitted, whereas viruses with the same basic specificity that did not have HA binding specificity to “long” α2,6, were not.
The present paper reports the following investigation:
“We have compared directly, by carbohydrate microarray analysis, the receptor-binding characteristics of two isolates of the novel pandemic H1N1 virus, Cal/09 and A/Hamburg/5/2009 (Ham/09), with those of a seasonal human H1N1 virus, A/Memphis/14/96-M (Mem/96), as representative of a virus well adapted to humans [and a reassortant human H3N2 virus A/Aichi/2/68 x PR8 (X31)]. As the HA of the novel H1N1 pandemic virus originated from a virus similar to triple reassortant swine H1N1 viruses, we compared one such example, A/Iowa/1/2006 (Iowa/06), isolated from a human infection, and an older close relative of classical swine H1N1 viruses, A/New Jersey/76 (NJ/76), the human isolate that initiated the concern of a pandemic threat in 1976.”
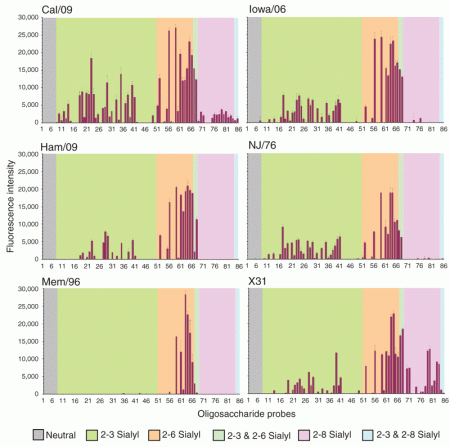
This is a really comprehensive analysis – for such a short communication – which throws up a number of interesting points. First, I was not aware it was possible to do “carbohydrate microarrays”! Second, the paper shows quite conclusively that the swine-derived AH1N1 viruses have a significantly wider range of receptor specificities than a standard seasonal AH1N1 virus, and – but to a lesser extent – than the reassortant H3N2 virus X31.
Carbohydrate microarray analyses of the six viruses investigated.
From the following article (with permission from NBT):
Receptor-binding specificity of pandemic influenza A (H1N1) 2009 virus determined by carbohydrate microarray.
Robert A Childs, Angelina S Palma, Steve Wharton, Tatyana Matrosovich, Yan Liu, Wengang Chai, Maria A Campanero-Rhodes, Yibing Zhang, Markus Eickmann, Makoto Kiso, Alan Hay, Mikhail Matrosovich & Ten Feizi.
Nature Biotechnology 27, 797 – 799 (2009).
doi:10.1038/nbt0909-797
Legend:
Numerical scores for the binding signals are shown as means of duplicate spots at 5 fmol per spot (with error bars). The microarrays consisted of eighty sialylated and six neutral lipid-linked oligosaccharide probes, printed on nitrocellulose-coated glass slides. These are listed in Supplementary Table 1 and arranged according to sialic acid linkage, oligosaccharide backbone chain length and sequence. The various types of terminal sialic acid linkage are indicated by the colored panels as defined at the bottom of the figure.
And what does all this mean, exactly? The authors sum it up well:
These results indicate that no major change in receptor-binding specificity of the HA was required for the emergent pandemic virus to acquire human-like characteristics and become established in the human population. …
The broader specificity, namely, the ability to bind to 2-3- in addition to 2-6-linked receptors is also pertinent to the greater virulence of the pandemic virus than seasonal influenza viruses observed in animal models, and its capacity to cause severe and fatal disease in humans, despite the generally mild nature of most infections. Binding to 2-3-linked receptors is thought to be associated with the ability of influenza viruses to infect the lower respiratory tract where there is a greater proportion of 2-3- relative to 2-6-linked sialyl glycans, although long chain 2-3-linked sialyl (poly-N-acetyllactosamine) sequences are present in ciliated bronchial epithelial cells in humans where they are the receptors for another human pathogen, Mycoplasma pneumoniae.
So there you have it: the viruses can get deeper in to your lungs than the standard flu – which, if it happens, can make you seriously ill.
So what happens if it gets better at binding the 2,3-type receptors in humans? Well, we’re only in the middle of the pandemic. We may yet find out the hard way.